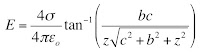One valuable skill in physics is the ability to make order of magnitude estimates, meaning to calculate something approximately right.Our first four homework problems at the end of Chapter 1 ask the student to estimate. These exercises are examples of Fermi Problems, after physicist Enrico Fermi who was a master of the skill.
Let us try another of these problems, based on some of the concepts from Chapter 15 about the natural background radiation dose. To be specific, let’s estimate the annual background dose from the radioactive isotope potassium-40 inside our bodies.
Problem 4 ½ Estimate the annual background dose (in mSv/year) from potassium-40 in our bodies. Look up or guess any information you need for this calculation, and clearly explain any assumptions you make.Begin by considering a single cell. Cells are about 10 microns in size, so their volume is about (10−5 m)3 = 10−15 m3 (the volume we use is not important; it will cancel out in the end). In nerves, the intracellular potassium ion concentration is about 100 mM, but this may overestimate the amount of potassium inside all types of cells. Moreover, the concentration of potassium ions in the extracellular space is much less that in the intracellular space. Let’s guess 50 mM for the average concentration of potassium in our body, meaning 50 millimoles/liter, or 50 moles/m3. If we multiply by Avogadro’s number (6 x 1023 molecules/mole), we get about 3 x 1025 molecules/m3. So, the number of potassium atoms per cell is 3 x 1010. This intermediate result is already interesting; there are over ten billion potassium ions in just one cell.
The 40K isotope is radioactive. Its abundance is about 0.01% (abundance data can be found in any table of the isotopes, or even by looking at wikipedia; I don’t know how you could guess that value from first principles). So, this means there are 3 x 106 40K atoms/cell (a little over a million). How rapidly do these decay? 40K has a half life of 1.25 x 109 years (again, see wikipedia), implying a decay rate of 0.693/1.25 x 109 = 5.5 x 10−10 decays per atom per year. Multiplying by the number of atoms/cell, we get 0.0017 decays per cell per year. This is another interesting result: an average cell has less than a one-percent chance of experiencing a 40K decay in a year. But we have a lot of cells (Russ and I estimate 2 x 1014 in IPMB), so your body suffers from about 3.4 x 1011 decays per year, or about ten thousand per second. (According to Wikipedia, this estimate is a factor of two too high, but we are not much worried about factors of two in such order-of-magnitude Fermi problems.)
How much energy does each decay deposit in our tissue? Beta decay accounts for 90% of all decays of 40K, and each decay releases an energy of about 1.3 MeV (decay energy data is a little harder to find, but appears in any good table of the isotopes; if you had merely guessed that 1 MeV is the order of magnitude of nuclear decay energies, you would not be too far off). Some of that energy goes to a neutrino, which leaves the body. Let’s assume that on average about 30% of the beta decay energy goes to the electron and that no electrons escape the body, so each decay deposits 0.4 MeV into the cell, or 6 x 10−14 joules. A gray (the unit of dose) is a joule per kilogram, so to calculate dose we need the mass of a cell, which to a first approximation is the product of the density of water (103 kg/m3) times the volume of the cell, or 10−12 kg (I told you the volume of the cell would cancel out). So, the dose is the number of decays per year (0.0017) times the energy per decay (6 x 10−14 J) divided by the mass (10−12 kg), or 0.0001 gray/year. Another unit of dose is the sievert, which accounts for biological damage in addition to energy deposition. A gray and a sievert are the same for electrons, so the annual background dose is 0.0001 Sv, or 0.1 mSv.
In Table 16.6 of IPMB, Russ and I estimate the annual background dose from all internal sources is about 0.3 mSv. Because 40K is not the only isotope in our body that is decaying (for example, carbon-14 is another), we seem to have gotten our order-of-magnitude estimate pretty close. One goal of Chapters 16 and 17 in IPMB is to refine such calculations. For medical purposes you need more accuracy; for a Fermi problem we did okay.