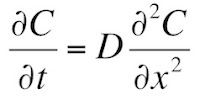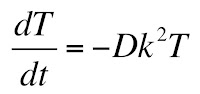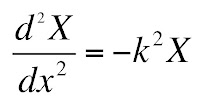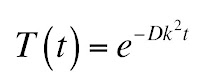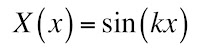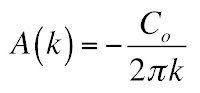In Chapter 4 of the 5th edition of Intermediate Physics for Medicine and Biology, Russ Hobbie and I introduce the diffusion equation (Equation 4.26)
where C is the concentration and D is the diffusion constant. We then study one-dimensional diffusion where initially (t = 0) the region to the left of x = 0 has a concentration Co, and the region to the right has a concentration of zero (Section 4.13). We show that the solution to the diffusion equation is (Equation 4.75)
where erf is the error function.
Some students don’t like error functions (really?). Moreover, often we can gain insight by solving a problem in several ways, obtaining solutions that are seemingly different yet equivalent. Let’s see if we can solve this problem in another way and avoid those pesky error functions. We will use a standard method for solving partial differential equations: separation of variables. Before I start, let’s agree to solve a slightly different problem: initially C(x,0) is Co/2 for x less than zero, and −Co/2 for x greater than zero. I do this so the solution will be odd in x: C(x,t) = −C(−x,t). At the end we can add the constant Co/2 and get back to our original problem. Now, let’s begin.
Assume the solution can be written as the product of a function of only t and a function of only x: C(x,t) = T(t) X(x). Plug this into the diffusion equation, simplify, and divide both sides by TX
The only way that the left and right hand sides can be equal at all values of x and t is if both are equal to a constant, which I will call −k2. This gives two ordinary differential equations
and
The solution to the first equation is an exponential
and the solution to the second is a sine
There is no cosine term because of the odd symmetry. Unfortunately, we don’t know the value of k. In fact, our solution can be a superposition of infinitely many values of k
where A(k) specifies the weighting.
To determine A(k), use the Fourier techniques developed in Chapter 11. The result is
How did I get that? Let me outline the process, leaving you to fill in the missing steps. I should warn you that a mathematician would worry about the convergence of the integrals we evaluate, but you and I’ll brush those concerns under the rug.
At t = 0, our solution becomes
Except for a missing factor of 2π, this looks just like the Fourier transform from Section 11.9 of IPMB. Next, multiply each side of the equation by sin(ωx), and integrate over all x. Then, use Equation 11.66b to express the integral of the product of sines as a delta function. You get
Both C(x,0) and sin(kx) are odd, so their product is even, and for x greater than zero C(x,0) is −Co/2. Therefore,
You know how to integrate sine (I hope you do!), so
Here is where things get dicey. We don’t know what cosine equals at infinity, but if we say it averages to zero the first term goes away and we get our result
Plugging in this expression for A(k) gives our solution for C(x,t). If we want to go back to our original problem with an initial condition of Co on the left and zero on the right, we must add Co/2. Thus
Let’s compare this solution with the one in Equation 4.75 (given above). Our new solution does not contain the error function! Those of you who dislike that function can celebrate. Unfortunately, we traded the error function for an integral that we can’t evaluate in closed form. So, you can have a function that you may be unfamiliar with and that has a funny name, or you can have an expression with common functions like the sine and the exponential inside an integral. Pick your poison.
Friday, July 24, 2015
Friday, July 17, 2015
Boston
 |
| Boston, by Boston. |
I was in high school in 1976 when I bought Boston’s famous debut album. That was a big year: it was the bicentennial of the United States, Jimmy Carter was elected president, Nadia Comaneci was earning 10s and then-Bruce Jenner won the decathlon in the Olympic games, much to my chagrin the Cincinnati Reds won the world series (but it wasn’t quite as exciting a series as the year before, which was the best world series ever), and the Apple Computer Company was formed by Steve Jobs and Steve Wozniak. My family moved from Fort Wayne, Indiana to Ashland, Ohio, and I spent the year playing tuba in the high school band, managing the high school baseball team, reading my first Isaac Asimov book, wondering if I should study physics in college, and listening to Chicago, the Eagles, Peter Frampton, the Wings, and Boston. The severe winter of 1976-1977 in northern Ohio and the simultaneous energy crisis resulted in my high school missing several weeks of classes, so some of my friends and I had time to establish our own garage band: “Hades.” We didn’t have a singer, and my role was to pick out the melody on an electric keyboard while the guitars and drums banged out behind me. Only a few years later disco music and the Bee Gees drove me from rock to country music, which I have listened to ever since.
My ears are still tingling a bit from the concert. How loud was it? Chapter 13 in the 5th edition of Intermediate Physics for Medicine and Biology discusses the decibel scale for measuring sound intensity, a logarithmic scale defined as log10(I/Io), where the sound intensity is
-->I and the reference Io is the minimum perceptible sound (10−12 W m−2). Table 13.1 in IPMB says 120 dB is the threshold for pain, and 130 dB is typical for the peak sound at a rock concert. Stephanie and I were sitting in the back of the amphitheater, so I doubt we ever experienced 130 dB, but we were up there pretty high on the decibel scale. I probably didn’t lose any hair cells in my cochlea (see Section 13.5), but I wonder how the band plays concerts night after night without suffering hearing loss. As people age, they tend to lose the ability to hear high tones: presbycusis. I may not have heard the music last night in the same way I heard it in 1976; some of those frequencies may be lost to me forever.
The leader of Boston is Tom Scholz, their 68-year-old guitar player and keyboardist. Scholz was educated as a mechanical engineer, and is one of the few rock musicians who might enjoy reading IPMB. I found that I could identify with Scholz in some respects: he is past his prime, no longer topping the charts or breaking new ground in rock and roll. But, after decades in the business, he’s still out there performing, playing his music, and even sometimes writing new songs. It makes me want to go write another paper!
 |
| My daughter Stephanie and I at a Boston concert at Freedom Hill Amphitheater in Sterling Heights, Michigan. |
Friday, July 10, 2015
The Machinery of Life
 |
| The Machinery of Life, by David Goodsell. |
In biology and medicine, we study objects that span a wide range of sizes: from giant redwood trees to individual molecules. Therefore, we begin with a brief discussion of length scales.At the end of this section, we conclude
To examine the relative sizes of objects in more detail, see Morrison et al. (1994) or Goodsell (2009).I have talked about the book Powers of Ten by Morrison et al. previously in this blog. I have also mentioned David Goodsell’s book The Machinery of Life several times, but until today I have never devoted an entire blog entry to it.
In the 4th edition of IPMB, Russ and I cited the first edition of The Machinery of Life (1998), and that is the edition that sits on my bookshelf. When preparing the 5th edition, we updated our references, so we now cite the second edition of Goodsell's book (2009). Is there much difference between the two? Yes! Like when Dorothy left Kansas to enter Oz, the first edition is all black and white but the second edition is in glorious color. And what a difference color makes in a book that is first and foremost visual. The second edition of The Machinery of Life is stunningly beautiful. It is not just a colorized version of the first edition; it is a whole new book. Goodsell writes in the preface
I created the illustrations in this book to help bridge this gulf and allow us to see the molecular structure of cells, if not directly, then in an artistic rendition. I have included two types of illustrations with this goal in mind: watercolor paintings which magnify a small portion of a living cell by one million times, showing the arrangement of molecules inside, and computer-generated pictures, which show the atomic details of individual molecules. In this second edition of The Machinery of Life, these illustrations are presented in full color, and they incorporate many of the exciting scientific advances of the 15 years since the first edition.I have often wondered how much molecular biology a biological or medical physicist needs to know. I suppose it depends on their research specialty, but in general I believe a physicist who has read and understood The Machinery of Life has most of what you need to begin working at the interface of physics and biology: An understanding of the relative scale of biological objects, an overview of the different types of biological molecules and their structures and functions, and a visual sense of how these molecules fit together to form a cell. To the physicist wanting an introduction to biology on the molecular scale, I recommend starting with The Machinery of Life. That’s why it was included in my ideal bookshelf.
As with the first edition, I have used several themes to tie the pictures together. One is that of scale. Most of us do not have a good concept of the relative sizes of water molecules, proteins, ribosomes, bacteria, and people. To assist with this understanding, I have drawn the illustrations at a few consistent magnifications. The views showing the interiors of living cells, as in the Frontispiece and scattered through the last half of the book, are all drawn at one million times magnification. Because of this consistent scale, you can flip between pages in these chapters and compare the sizes of DNA, lipid membranes, nuclear pores, and all of the other molecular machinery of living cells. The computer-generated figures of individual molecules are also drawn at a few consistent scales to allow easy comparison.
I have also drawn the illustrations using a consistent style, again to allow easy comparison. A space-filling representation that shows each atom as a sphere is used for all the illustrations of molecules. The shapes of the molecules in the cellular pictures are simplified versions of these space-filling pictures, capturing the overall form of the molecule without showing the location of every atom. The colors, of course, are completely arbitrary since most of these molecules are colorless. I have chosen them to highlight the functional features of the molecules and cellular environments.
Goodsell fans might enjoy visiting his website: http://mgl.scripps.edu/people/goodsell. There you can download a beautiful poster of different proteins, all drawn to scale. There are many other illustrations and publications. Enjoy!
Friday, July 3, 2015
Fermi Problems and the Annual Background Radiation Dose from Potassium-40
In the very first section of the 5th edition of Intermediate Physics for Medicine and Biology, Russ Hobbie and I emphasize the importance of being able to estimate.
Let us try another of these problems, based on some of the concepts from Chapter 15 about the natural background radiation dose. To be specific, let’s estimate the annual background dose from the radioactive isotope potassium-40 inside our bodies.
The 40K isotope is radioactive. Its abundance is about 0.01% (abundance data can be found in any table of the isotopes, or even by looking at wikipedia; I don’t know how you could guess that value from first principles). So, this means there are 3 x 106 40K atoms/cell (a little over a million). How rapidly do these decay? 40K has a half life of 1.25 x 109 years (again, see wikipedia), implying a decay rate of 0.693/1.25 x 109 = 5.5 x 10−10 decays per atom per year. Multiplying by the number of atoms/cell, we get 0.0017 decays per cell per year. This is another interesting result: an average cell has less than a one-percent chance of experiencing a 40K decay in a year. But we have a lot of cells (Russ and I estimate 2 x 1014 in IPMB), so your body suffers from about 3.4 x 1011 decays per year, or about ten thousand per second. (According to Wikipedia, this estimate is a factor of two too high, but we are not much worried about factors of two in such order-of-magnitude Fermi problems.)
How much energy does each decay deposit in our tissue? Beta decay accounts for 90% of all decays of 40K, and each decay releases an energy of about 1.3 MeV (decay energy data is a little harder to find, but appears in any good table of the isotopes; if you had merely guessed that 1 MeV is the order of magnitude of nuclear decay energies, you would not be too far off). Some of that energy goes to a neutrino, which leaves the body. Let’s assume that on average about 30% of the beta decay energy goes to the electron and that no electrons escape the body, so each decay deposits 0.4 MeV into the cell, or 6 x 10−14 joules. A gray (the unit of dose) is a joule per kilogram, so to calculate dose we need the mass of a cell, which to a first approximation is the product of the density of water (103 kg/m3) times the volume of the cell, or 10−12 kg (I told you the volume of the cell would cancel out). So, the dose is the number of decays per year (0.0017) times the energy per decay (6 x 10−14 J) divided by the mass (10−12 kg), or 0.0001 gray/year. Another unit of dose is the sievert, which accounts for biological damage in addition to energy deposition. A gray and a sievert are the same for electrons, so the annual background dose is 0.0001 Sv, or 0.1 mSv.
In Table 16.6 of IPMB, Russ and I estimate the annual background dose from all internal sources is about 0.3 mSv. Because 40K is not the only isotope in our body that is decaying (for example, carbon-14 is another), we seem to have gotten our order-of-magnitude estimate pretty close. One goal of Chapters 16 and 17 in IPMB is to refine such calculations. For medical purposes you need more accuracy; for a Fermi problem we did okay.
One valuable skill in physics is the ability to make order of magnitude estimates, meaning to calculate something approximately right.Our first four homework problems at the end of Chapter 1 ask the student to estimate. These exercises are examples of Fermi Problems, after physicist Enrico Fermi who was a master of the skill.
Let us try another of these problems, based on some of the concepts from Chapter 15 about the natural background radiation dose. To be specific, let’s estimate the annual background dose from the radioactive isotope potassium-40 inside our bodies.
Problem 4 ½ Estimate the annual background dose (in mSv/year) from potassium-40 in our bodies. Look up or guess any information you need for this calculation, and clearly explain any assumptions you make.Begin by considering a single cell. Cells are about 10 microns in size, so their volume is about (10−5 m)3 = 10−15 m3 (the volume we use is not important; it will cancel out in the end). In nerves, the intracellular potassium ion concentration is about 100 mM, but this may overestimate the amount of potassium inside all types of cells. Moreover, the concentration of potassium ions in the extracellular space is much less that in the intracellular space. Let’s guess 50 mM for the average concentration of potassium in our body, meaning 50 millimoles/liter, or 50 moles/m3. If we multiply by Avogadro’s number (6 x 1023 molecules/mole), we get about 3 x 1025 molecules/m3. So, the number of potassium atoms per cell is 3 x 1010. This intermediate result is already interesting; there are over ten billion potassium ions in just one cell.
The 40K isotope is radioactive. Its abundance is about 0.01% (abundance data can be found in any table of the isotopes, or even by looking at wikipedia; I don’t know how you could guess that value from first principles). So, this means there are 3 x 106 40K atoms/cell (a little over a million). How rapidly do these decay? 40K has a half life of 1.25 x 109 years (again, see wikipedia), implying a decay rate of 0.693/1.25 x 109 = 5.5 x 10−10 decays per atom per year. Multiplying by the number of atoms/cell, we get 0.0017 decays per cell per year. This is another interesting result: an average cell has less than a one-percent chance of experiencing a 40K decay in a year. But we have a lot of cells (Russ and I estimate 2 x 1014 in IPMB), so your body suffers from about 3.4 x 1011 decays per year, or about ten thousand per second. (According to Wikipedia, this estimate is a factor of two too high, but we are not much worried about factors of two in such order-of-magnitude Fermi problems.)
How much energy does each decay deposit in our tissue? Beta decay accounts for 90% of all decays of 40K, and each decay releases an energy of about 1.3 MeV (decay energy data is a little harder to find, but appears in any good table of the isotopes; if you had merely guessed that 1 MeV is the order of magnitude of nuclear decay energies, you would not be too far off). Some of that energy goes to a neutrino, which leaves the body. Let’s assume that on average about 30% of the beta decay energy goes to the electron and that no electrons escape the body, so each decay deposits 0.4 MeV into the cell, or 6 x 10−14 joules. A gray (the unit of dose) is a joule per kilogram, so to calculate dose we need the mass of a cell, which to a first approximation is the product of the density of water (103 kg/m3) times the volume of the cell, or 10−12 kg (I told you the volume of the cell would cancel out). So, the dose is the number of decays per year (0.0017) times the energy per decay (6 x 10−14 J) divided by the mass (10−12 kg), or 0.0001 gray/year. Another unit of dose is the sievert, which accounts for biological damage in addition to energy deposition. A gray and a sievert are the same for electrons, so the annual background dose is 0.0001 Sv, or 0.1 mSv.
In Table 16.6 of IPMB, Russ and I estimate the annual background dose from all internal sources is about 0.3 mSv. Because 40K is not the only isotope in our body that is decaying (for example, carbon-14 is another), we seem to have gotten our order-of-magnitude estimate pretty close. One goal of Chapters 16 and 17 in IPMB is to refine such calculations. For medical purposes you need more accuracy; for a Fermi problem we did okay.
Friday, June 26, 2015
The Electric Potential of a Rectangular Sheet of Charge - Resolved
In the January 9 post of this blog, I challenged readers to find the electrical potential V(z) that will give you the electric field E(z) of Eq. 6.10 in the 5th edition of Intermediate Physics for Medicine and Biology
In other words, the goal is to find V(z) such that E = − dV/dz produces Eq. 6.10. In the comments at the bottom of the post, a genius named Adam Taylor made a suggestion for V(z) (I love it when people leave comments in this blog). When I tried his expression for the potential, it almost worked, but not quite (of course, there is always a chance I have made a mistake, so check it yourself). But I was able to fix it up with a slight modification. I now present to you, dear reader, the potential:
How do you interpret this ugly beast? The key is the last term, z times the inverse tangent. When you take the z derivative of V(z), you must use the product rule on this term. One derivative in the product rule eliminates the leading z and gives you exactly the inverse tangent you need in the expression for the electric field. The other gives z times a derivative of the inverse tangent, which is complicated. The two terms containing the logarithms are needed to cancel the mess that arises from differentiating tan−1.
I don’t know what there is to gain from having this expression for the potential, but somehow it comforts me to know that if there is an analytic equation for E there is also an analytic equation for V.
In other words, the goal is to find V(z) such that E = − dV/dz produces Eq. 6.10. In the comments at the bottom of the post, a genius named Adam Taylor made a suggestion for V(z) (I love it when people leave comments in this blog). When I tried his expression for the potential, it almost worked, but not quite (of course, there is always a chance I have made a mistake, so check it yourself). But I was able to fix it up with a slight modification. I now present to you, dear reader, the potential:
How do you interpret this ugly beast? The key is the last term, z times the inverse tangent. When you take the z derivative of V(z), you must use the product rule on this term. One derivative in the product rule eliminates the leading z and gives you exactly the inverse tangent you need in the expression for the electric field. The other gives z times a derivative of the inverse tangent, which is complicated. The two terms containing the logarithms are needed to cancel the mess that arises from differentiating tan−1.
I don’t know what there is to gain from having this expression for the potential, but somehow it comforts me to know that if there is an analytic equation for E there is also an analytic equation for V.
Friday, June 19, 2015
Dr. Euler’s Fabulous Formula Cures Many Mathematical Ills
 |
| Dr. Euler's Fabulous Formula, by Paul Nahin. |
eiθ = cosθ + i sinθ .
I liked the book, in part because Nahin and I seem to have similar tastes: we both favor the illustrations of Norman Rockwell over the paintings of Jackson Pollock, we both like to quote Winston Churchill, and we both love limericks:
I used to think math was no fun,Nahin’s book contains a lot of math, and I admit I didn’t go through it all in detail. A large chunk of the text talks about the Fourier series, which Russ Hobbie and I develop in Chapter 11 of IPMB. Nahin motivates the study of the Fourier series as a tool to solve the wave equation. We discuss the wave equation in Chapter 13 of IPMB, but never make the connection between the Fourier series and this equation, perhaps because biomedical applications don’t rely on such an analysis as heavily as, say, predicting how a plucked string vibrates.
‘Cause I couldn’t see how it was done.
Now Euler’s my hero
For I now see why zero,
Equals eπi + 1.
Nahin delights in showing how interesting mathematical relationships arise from Fourier analysis. I will provide one example, closely related to a calculation in IPMB. In Section 11.5, we show that the Fourier series of the square wave (y(t) = 1 for t from 0 to T/2 and equal to -1 for t from T/2 to T) is
y(t) = Σ bk cos(k2πt/T)
where the sum is over all odd values of k (k = 1, 3, 5, ....) and bk = 4/(π k). Evaluate both expressions for y(t) at t = T/4. You get
π/4 = 1 – 1/3 + 1/5 – 1/7 +…
This lovely result is hidden in IPMB’s Eq. 11.36. Warning: this is not a particularly useful algorithm for calculating π, as it converges slowly; including ten terms in the sum gives π = 3.04, which is still over 3% off.
In Figure 11.17, Russ and I discuss the Gibbs phenomenon: spikes that occur in y(t) at discontinuities when the Fourier series includes only a finite number of terms. Nahin makes the same point with the periodic function y(t) = (π – t)/2 for t from 0 to 2π. He describes the history of the Gibbs phenomena, which arises from a series of published letters between Josiah Gibbs, Albert Michelson, A. E. H. Love, and Henri Poincare. Interestingly, the Gibbs phenomenon was discovered long before Gibbs by the English mathematician Henry Wilbraham.
Fourier series did not originate with Joseph Fourier. Euler, for example, was known to write such trigonometric series. Fourier transforms (the extension of Fourier series to nonperiodic functions), on the other hand, were first presented by Fourier. Nahin discusses many of the same topics that Russ and I cover, including the Dirac delta function, Parseval’s theorem, convolutions, and the autocorrelation.
Nahin concludes with a section about Euler the man and mathematical physicist. I found an interesting connection to biology and medicine: when hired in 1727 by the Imperial Russian Academy of Sciences, it was as a professor of physiology. Euler spent several months before he left for Russia studying physiology, so he would not be totally ignorant of the subject when he arrived in Saint Petersburg!
I will end with a funny story of my own. I was working at Vanderbilt University just as Nashville was enticing a professional football team to move there. One franchise that was looking to move was the Houston Oilers. Once the deal was done, folks in Nashville began debating what to call their new team. They wanted a name that would respect the team’s history, but would also be fitting for its new home. Nashville has always prided itself as the home of many colleges and universities, so a name out of academia seemed appropriate. Some professors in Vanderbilt’s Department of Mathematics came up with what I thought was the perfect choice: call the team the Nashville Eulers. Alas, the name didn’t catch on, but at least I never again was uncertain about how to pronounce Euler.
Friday, June 12, 2015
Circularly Polarized Excitation Pulses, Spin Lock, and T1ρ
In Chapter 18 of the 5th edition of Intermediate Physics for Medicine and Biology, Russ Hobbie and I discuss magnetic resonance imaging. We describe how spins precess in a magnetic field, Bo = Bo k, at their Larmor frequency ω, and show that this behavior is particularly simple when expressed in a rotating frame of reference. We then examine radio-frequency excitation pulses by adding an oscillating magnetic field. Again, the analysis is simpler in the rotating frame. In our book, we apply the oscillating field in the laboratory frame’s x-direction, B1 = B1 cos(ωt) i. The equations of the magnetization are complicated in the rotating frame (Eqs. 18.22-18.24), but become simpler when we average over time (Eq. 18.25). The time-averaged magnetization rotates about the rotating frame’s x' axis with angular frequency γB1/2, where γ is the spin’s gyromagnetic ratio. This motion is crucial for exciting the spins; it rotates them from their equilibrium position parallel to the static magnetic field Bo into a plane perpendicular to Bo.
When I teach medical physics (PHY 326 here at Oakland University), I go over this derivation in class, but the students still need practice. I have them analyze some related examples as homework. For instance, the oscillating magnetic field can be in the y direction, B1 = B1 cos(ωt) j, or can be shifted in time, B1 = B1 sin(ωt) i. Sometimes I even ask them to analyze what happens when the oscillating magnetic field is in the z direction, B1 = B1 cos(ωt) k, parallel to the static field. This orientation is useless for exciting spins, but is useful as practice.
Yet another way to excite spins is using a circularly polarized magnetic field, B1 = B1 cos(ωt) i – B1 sin(ωt) j. The analysis of this case is similar to the one in IPMB, with one twist: you don’t need to average over time! Below is a new homework problem illustrating this.
Now let’s assume that an RF excitation pulse rotates the magnetization so it aligns with the x' rotating axis. Once the pulse ends, what happens? Well, nothing happens unless we account for relaxation. Without relaxation the magnetization precesses around the static field, which means it just sits there stationary in the rotating frame. But we know that relaxation occurs. Consider the mechanism of dephasing, which underlies the T2* relaxation time constant. Slight heterogeneities in Bo mean that different spins precess at different Larmor frequencies, causing the spins to lose phase coherence, decreasing the net magnetization.
Next, consider the case of spin lock. Imagine an RF pulse rotates the magnetization so it is parallel to the x' rotating axis. Then, when the excitation pulse is over, immediately apply a circularly polarized RF pulse at the Larmor frequency, called B2, which is aligned along the x' rotating axis. In the rotating frame the magnetization is parallel to B2, so nothing happens. Why bother? Consider those slight heterogeneities in Bo that led to T2* relaxation. They will cause the spins to dephase, picking up a component in the y' direction. But a component along y' will start to precess around B2. Rather than dephasing, B2 causes the spins to wobble around in the rotating frame, precessing about x', with no net tendency to dephase. You just killed the mechanism leading to T2*! Wow!
Will the spins just precess about B2 forever? No, eventually other mechanisms will cause them to relax toward their equilibrium value. Their time constant will not be T1 or T2 or even T2*, but something else called T1ρ. Typcially, T1ρ is much longer than T2*. To measure T1ρ, apply a 90 degree excitation pulse, then apply a RF spin lock oscillation and record the free induction decay. Fit the decay to an exponential, and the time constant you obtain is T1ρ. (I am not a MRI expert: I am not sure how you can measure a free induction decay when a spin lock field is present. I would think the spin lock field would swamp the FID.)
T1ρ is sometimes measured to learn about the structure of cartilage. It is analogous to T1 relaxation in the laboratory frame, which explains its name. Because B2 is typically much weaker than Bo, T1ρ is sensitive to a different range of correlation times than T1 or T2 (see Fig. 18.12 in IPMB).
When I teach medical physics (PHY 326 here at Oakland University), I go over this derivation in class, but the students still need practice. I have them analyze some related examples as homework. For instance, the oscillating magnetic field can be in the y direction, B1 = B1 cos(ωt) j, or can be shifted in time, B1 = B1 sin(ωt) i. Sometimes I even ask them to analyze what happens when the oscillating magnetic field is in the z direction, B1 = B1 cos(ωt) k, parallel to the static field. This orientation is useless for exciting spins, but is useful as practice.
Yet another way to excite spins is using a circularly polarized magnetic field, B1 = B1 cos(ωt) i – B1 sin(ωt) j. The analysis of this case is similar to the one in IPMB, with one twist: you don’t need to average over time! Below is a new homework problem illustrating this.
Problem 13 1/2. Assume you have a static magnetic field in the z direction and an oscillating, circularly polarized magnetic field in the x-y plane, B =Bo k + B1 cos(ωt) i – B1 sin(ωt) j.
a) Use Eq. 18.12 to derive the equations for the magnetization M in the laboratory frame of reference (ignore relaxation).
b) Use Eq. 18.18 to transform to the rotating coordinate system and derive equations for M'.
c) Interpret these results physically.I get the same equations as derived in IPMB (Eq. 18.25) except for a factor of one half; the angular frequency in the rotating frame is ω1 = γ B1. Not having to average over time makes the result easier to visualize. You don’t get a complex motion that—on average—rotates the magnetization. Instead, you get a plain old rotation. You can understand this behavior qualitatively without any math by realizing that in the rotating coordinate system the RF circularly polarized magnetic field is stationary, pointing in the x’ direction. The spins simply precess around the seemingly static B1'= B1 i', just like the spins precess around the static Bo = Bo k in the laboratory frame.
Now let’s assume that an RF excitation pulse rotates the magnetization so it aligns with the x' rotating axis. Once the pulse ends, what happens? Well, nothing happens unless we account for relaxation. Without relaxation the magnetization precesses around the static field, which means it just sits there stationary in the rotating frame. But we know that relaxation occurs. Consider the mechanism of dephasing, which underlies the T2* relaxation time constant. Slight heterogeneities in Bo mean that different spins precess at different Larmor frequencies, causing the spins to lose phase coherence, decreasing the net magnetization.
Next, consider the case of spin lock. Imagine an RF pulse rotates the magnetization so it is parallel to the x' rotating axis. Then, when the excitation pulse is over, immediately apply a circularly polarized RF pulse at the Larmor frequency, called B2, which is aligned along the x' rotating axis. In the rotating frame the magnetization is parallel to B2, so nothing happens. Why bother? Consider those slight heterogeneities in Bo that led to T2* relaxation. They will cause the spins to dephase, picking up a component in the y' direction. But a component along y' will start to precess around B2. Rather than dephasing, B2 causes the spins to wobble around in the rotating frame, precessing about x', with no net tendency to dephase. You just killed the mechanism leading to T2*! Wow!
Will the spins just precess about B2 forever? No, eventually other mechanisms will cause them to relax toward their equilibrium value. Their time constant will not be T1 or T2 or even T2*, but something else called T1ρ. Typcially, T1ρ is much longer than T2*. To measure T1ρ, apply a 90 degree excitation pulse, then apply a RF spin lock oscillation and record the free induction decay. Fit the decay to an exponential, and the time constant you obtain is T1ρ. (I am not a MRI expert: I am not sure how you can measure a free induction decay when a spin lock field is present. I would think the spin lock field would swamp the FID.)
T1ρ is sometimes measured to learn about the structure of cartilage. It is analogous to T1 relaxation in the laboratory frame, which explains its name. Because B2 is typically much weaker than Bo, T1ρ is sensitive to a different range of correlation times than T1 or T2 (see Fig. 18.12 in IPMB).
Friday, June 5, 2015
Robert Plonsey (1924-2015)
 |
| Bioelectricity: A Quantitative Approach, by Plonsey and Barr. |
Plonsey had an enormous impact on my research when I was in graduate school. For example, in 1968 John Clark and Plonsey calculated the intracellular and extracellular potentials produced by a propagating action potential along a nerve axon (“The Extracellular Potential Field of a Single Active Nerve Fiber in a Volume Conductor,” Biophysical Journal, Volume 8, Pages 842−864). Russ and I outline this calculation--which uses Bessel functions and Fourier transforms--in IPMB’s Homework Problem 30 of Chapter 6. In one of my first papers, Jim Woosley, my PhD advisor John Wikswo, and I extended Clark and Plonsey’s calculation to predict the axon’s magnetic field (Woosley, Roth, and Wikswo, 1985, “The Magnetic Field of a Single Axon: A Volume Conductor Model,” Mathematical Bioscience, Volume 76, Pages 1−36). I have described Clark and Plonsey’s groundbreaking work before in this blog.
I associate Plonsey most closely with the development of the bidomain model of cardiac tissue. The 1980s was an exciting time to be doing cardiac electrophysiology, and Duke University, where Plonsey worked, was the hub of this activity. Wikswo, Nestor Sepulveda, and I, all at Vanderbilt University, had to run fast to compete with the Duke juggernaut that included Plonsey, Barr, Ray Ideker, Theo Pilkington, and Madison Spach, as well as a triumvirate of then up-and-coming researchers from my generation: Natalia Trayanova, Wanda Krassowska, and Craig Henriquez. To get a glimpse of these times (to me, the “good old days”), read Henriquez’s “A Brief History of Tissue Models for Cardiac Electrophysiology” (IEEE Transaction on Biomedical Engineering, Volume 61, Pages 1457−1465) published last year.
My first work on the bidomain model was to extend Clark and Plonsey’s calculation of the potential along a nerve axon to an analogous calculation along a cylindrical strand of cardiac tissue, such as a papillary muscle (Roth and Wikswo, 1986, “A Bidomain Model for Extracellular Potential and Magnetic Field of Cardiac Tissue,” IEEE Transaction on Biomedical Engineering, Volume 33, Pages 467−469). I remember what an honor it was for me when Plonsey and Barr cited our paper (and mentioned John and me by name!) in their 1987 article “Interstitial Potentials and Their Change with Depth into Cardiac Tissue” (Biophysical Journal, Volume 51, Pages 547−555). That was heady stuff for a nobody graduate student who could count his citations on his ten fingers.
One day Wikswo returned from a conference and told us about a talk he heard, by either Plonsey or Barr (I don’t recall which), describing the action current distribution produced by a outwardly propagating wave front in a sheet of cardiac tissue (Plonsey and Barr, 1984, “Current Flow Patterns in Two-Dimensional Anisotropic Bisyncytia with Normal and Extreme Conductivities,” Biophysical Journal, Volume 45, Pages 557−571). Wikswo realized immediately that their calculations implied the wave front would have a distinctive magnetic signature, which he and Nestor Sepulveda reported in 1987 (“Electric and Magnetic Fields From Two-Dimensional Anisotropic Bisyncytia,” Biophysical Journal, Volume 51, Pages 557−568).
In another paper, Barr and Plonsey derived a numerical method to solve the bidomain equations including the nonlinear ion channel kinetics (Barr and Plonsey, 1984, “Propagation of Excitation in Idealized Anisotropic Two-Dimensional Tissue,” Biophysical Journal, Volume 45, Pages 1191−1202). This paper was the inspiration for my own numerical algorithm (Roth, 1991, “Action Potential Propagation in a Thick Strand of Cardiac Muscle,” Circulation Research, Volume 68, Pages 162−173). In my paper, I cited several of Plonsey’s articles, including one by Plonsey, Henriquez, and Trayanova about an “Extracellular (Volume Conductor) Effect on Adjoining Cardiac Muscle Electrophysiology” (1988, Medical and Biological Engineering and Computing, Volume 26, Pages 126−129), which shared the conclusion I reached that an adjacent bath can dramatically affect action potential propagation in cardiac tissue. Indeed, Henriquez (Plonsey’s graduate student) and Plonsey were following a similar line of research, resulting in two papers partially anticipating mine (Henriquez and Plonsey, 1990, “Simulation of Propagation Along a Cylindrical Bundle of Cardiac Tissue—I: Mathematical Formulation,” IEEE Transactions on Biomedical Engineering, Volume 37, Pages 850−860; and Henriquez and Plonsey, 1990, “Simulation of Propagation Along a Cylindrical Bundle of Cardiac Tissue—II: Results of Simulation,” IEEE Transactions on Biomedical Engineering, Volume 37, Pages 861−875.)
In parallel with this research, Ideker was analyzing how defibrillation shocks affected cardiac tissue, and in 1986 Plonsey and Barr published two papers presenting their saw tooth model (“Effect of Microscopic and Macroscopic Discontinuities on the Response of Cardiac Tissue to Defibrillating (Stimulating) Currents,” Medical and Biological Engineering and Computing, Volume 24, Pages 130−136; “Inclusion of Junction Elements in a Linear Cardiac Model Through Secondary Sources: Application to Defibrillation,” Volume 24, Pages 127−144). (It’s interesting how many of Plonsey’s papers were published as pairs.) I suspect that if in 1989 Sepulveda, Wikswo and I had not published our article about unipolar stimulation of cardiac tissue (“Current Injection into a Two-Dimensional Anisotropic Bidomain,” Biophysical Journal, Volume 55, Pages 987−999), one of the Duke researchers—perhaps Plonsey himself—would have soon performed the calculation. (To learn more about the Sepulveda et al paper, read my May 2009 blog entry.)
In January 1991 I visited Duke and gave a talk in the Emerging Cardiovascular Technologies Seminar Series, where I had the good fortune to meet with Plonsey. Somewhere I have a videotape of that talk; I suppose I should get it converted to a digital format. When I was working at the National Institutes of Health in the mid 1990s, Plonsey was a member of an external committee that assessed my work, as a sort of tenure review. I will always be grateful for the positive feedback I received, although it was to no avail because budget cuts and a hiring freeze led to my leaving NIH in 1995. Plonsey retired from Duke in 1996, and our paths didn’t cross again. He was a gracious gentleman who I will always have enormous respect for. Indeed, the first seven years of my professional life were spent traveling down a path parallel to and often intersecting his; to put it more aptly, I was dashing down a trail he had blazed.
Robert Plonsey was a World War Two veteran (we are losing them too fast these days), and a leader in establishing biomedical engineering as an academic discipline. You can read his obituary here and here.
I will miss him.
Friday, May 29, 2015
Taylor's Series
In Appendix D of the 5th edition of Intermediate Physics for Medicine and Biology, Russ Hobbie and I review Taylor’s Series. Our Figures D.3 and D.4 show better and better approximations to the exponential function, ex, found by using more and more terms of its Taylor’s series. As we add terms, the approximation improves for small |x| and diverges more slowly for large |x|. Taking additional terms from the Taylor’s series approximates the exponential by higher and higher order polynomials. This is all interesting and useful, but the exponential looks similar to a polynomial, at least for positive x, so it is not too surprising that polynomials do a decent job approximating the exponential.
A more challenging function to fit with a Taylor’s Series would look nothing like a polynomial, which always grows to plus or minus infinity at large |x|. I wonder how the Taylor’s Series does approximating a bounded function; perhaps a function that oscillates back and forth a lot? The natural choice is the sine function.
The Taylor’s Series of sin(x) is
The figure below shows the sine function and its various polynomial approximations.
The red curve is the sine function itself. The simplest approximation is simply sin(x) = x, which gives the yellow straight line. It looks good for |x| less than one, but quickly diverges from sine at large |x|. The green curve is sin(x) = x – x3/6. It rises to a maximum and then falls, much like sin(x), but heads off to plus or minus infinity relatively quickly. The cyan curve is sin(x) = x – x3/6 + x5/120. It captures the first peak of the oscillation well, but then fails. The royal blue curve is sin(x) = x – x3/6 + x5/120 – x7/5040. It is an excellent approximation of sin(x) out to x = π. The violet curve is sin(x) = x – x3/6 + x5/120 – x7/5040 + x9/362880. It begins to capture the second oscillation, but then diverges. You can see the Taylor’s Series is working hard to represent the sine function, but it is not easy.
Appendix D in IPMB gives a table of values of the exponential and its different Taylor’s Series approximations. Below I create a similar table for the sine. Because the sine and all its series approximations are odd functions, I only consider positive values of x.
One final thought. Russ and I title Appendix D as “Taylor’s Series” with an apostrophe s. Should we write “Taylor Series” instead, without the apostrophe s? Wikipedia just calls it the “Taylor Series.” I’ve seen it both ways, and I don’t know which is correct. Any opinions?
A more challenging function to fit with a Taylor’s Series would look nothing like a polynomial, which always grows to plus or minus infinity at large |x|. I wonder how the Taylor’s Series does approximating a bounded function; perhaps a function that oscillates back and forth a lot? The natural choice is the sine function.
The Taylor’s Series of sin(x) is
sin(x) = x – x3/6 + x5/120 – x7/5040 + x9/362880 - …
The figure below shows the sine function and its various polynomial approximations.
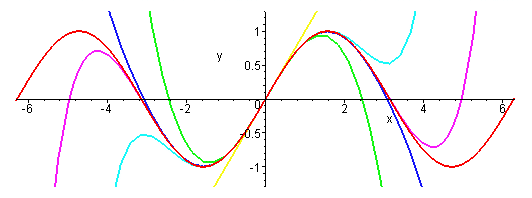 |
| The sine function and its various polynomial approximations, from: http://www.peterstone.name/Maplepgs/images/Maclaurin_sine.gif |
Appendix D in IPMB gives a table of values of the exponential and its different Taylor’s Series approximations. Below I create a similar table for the sine. Because the sine and all its series approximations are odd functions, I only consider positive values of x.
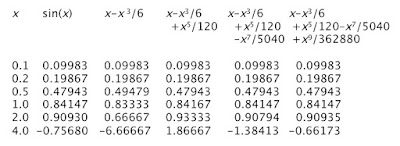 |
| A table of values for sin(x) and its various polynomial approximations. |
One final thought. Russ and I title Appendix D as “Taylor’s Series” with an apostrophe s. Should we write “Taylor Series” instead, without the apostrophe s? Wikipedia just calls it the “Taylor Series.” I’ve seen it both ways, and I don’t know which is correct. Any opinions?
Friday, May 22, 2015
Progress Toward a Deployable SQUID-Based Ultra-Low Field MRI System for Anatomical Imaging
When surfing the web, I like to visit medicalphysicsweb.org. This site, maintained by the Institute of Physics, always publishes interesting and up-to-date information about physics applied to medicine. Readers of the 5th Edition of Intermediate Physics for Medicine and Biology should visit it regularly.
A recent article discusses a paper by Michelle Espy and her colleagues about “Progress Toward a Deployable SQUID-Based Ultra-Low Field MRI System for Anatomical Imaging” (IEEE Transactions on Applied Superconductivity, Volume 25, Article Number 1601705, June 2015). The abstract is given below.
A magnetic field of 1500 mT is usually produced by a coil that must be kept at low temperatures to maintain the wire as a superconductor. A 100 mT field does not require superconductivity, but the needed current generates enough heat that the 510-turn copper coil needs to be cooled by liquid nitrogen, and even still the current in the coil must be turned off half the time (50% duty cycle) to avoid overheating.
Once the polarization field turns off, the spins precess with the Larmor frequency for a 0.2 mT magnetic field. The gyromagnetic ratio of protons is 42.6 kHz/mT, implying a Larmor frequency of 8.5 kHz, compared with 64,000 kHz for a clinical machine. So, the Larmor frequencies differ by a factor of 7500.
The magnetic resonance signal recorded in a clinical system is large compared to an ultra-low device because the magnetization is larger by a factor of 15 and the Larmor frequency is larger by a factor of 7500, implying a signal over a hundred thousand times larger. Espy et al. get around this problem by measuring the signal with a Superconducting Quantum Interference Device (SQUID) magnetometer, like those used in magnetoencephalography (see Section 8.9 in IPMB).
Preliminary experiments were performed in a heavy and expensive magnetically shielded room (again, like those used when measuring the MEG). However, second-order gradiometer pickup coils reduce the noise sufficiently that the shielded room is unnecessary.
To perform imaging, Espy et al. use magnetic field gradients of about 0.00025 mT/mm, compared with 0.01 mT/mm gradients typical for clinical MRI. For two objects 1 mm apart, a clinical imaging system would therefore produce a fractional frequency shift of 0.01/1500 = 0.0000067 or 6.7 ppm, whereas a low-field device has a fractional shift of 0.00025/0.2 = 0.00125 or 1250 ppm. Therefore, the clinical magnetic field needs to be extremely homogeneous (on the order of parts per million) to avoid artifacts, whereas a low-field device can function with heterogeneities hundreds of times larger.
The relaxation time constants for gray matter in the brain are reported by Espy et al. as about T1 = 600 ms and T2 = 80 ms. In clinical devices, the value of T1 is about half that, and T2 is about the same. Based on Figure 18.12 and Equation 18.35 in IPMB, I’m not surprised that T2 is largely independent of the magnetic field strength. However, in strong fields T1 increases as the square of the magnetic field (or the square of the Larmor frequency), so I was initially expecting a much smaller value of T1 for the low-field device. But once the Larmor frequency drops to values less than the typical correlation time of the spins, T1 becomes independent of the magnetic field strength (Equation 18.34 in IPMB). I assume that is what is happening here, and explains why T1 drops by only a factor of two when the magnetic field is reduced by a factor of 7500.
I find the differences between radio-frequency excitation pulses to be interesting. In clinical imaging, if the excitation pulse has a duration of about 1 ms and the Larmor frequency is 64,000 kHz, there are 64,000 oscillations of the radio-frequency magnetic field in a single π/2 pulse. Espy et al. used a 4 ms duration excitation pulse and a Larmor frequency of 8.5 kHz, implying just 34 oscillations per pulse. I have always worried that illustrations such as Figure 18.23 in IPMB mislead because they show the Larmor frequency as being not too different from the excitation pulse duration. For low-field MRI, however, this picture is realistic.
Does low-field MRI have advantages? You don’t need the heavy, expensive superconducting coil to generate a large static field, but you do need SQUID magnetometers to record the small signal, so you don’t avoid the need for cryogenics. The medicalphysicsweb article weighs the pros and cons. For instance, the power requirements for a low-field device are relatively small, and it is more portable, but the imaging times are long. The safety hazards caused by metal are much less in a low-field system, but the impact of stray magnetic fields is greater. I’m skeptical about the ultimate usefulness of ultra low-field MRI, but it’ll be fun to watch if Espy and her team can prove me wrong.
A recent article discusses a paper by Michelle Espy and her colleagues about “Progress Toward a Deployable SQUID-Based Ultra-Low Field MRI System for Anatomical Imaging” (IEEE Transactions on Applied Superconductivity, Volume 25, Article Number 1601705, June 2015). The abstract is given below.
Magnetic resonance imaging (MRI) is the best method for non-invasive imaging of soft tissue anatomy, saving countless lives each year. But conventional MRI relies on very high fixed strength magnetic fields, ≥ 1.5 T, with parts-per-million homogeneity, requiring large and expensive magnets. This is because in conventional Faraday-coil based systems the signal scales approximately with the square of the magnetic field. Recent demonstrations have shown that MRI can be performed at much lower magnetic fields (∼100 μT, the ULF regime). Through the use of pulsed prepolarization at magnetic fields from ∼10–100 mT and SQUID detection during readout (proton Larmor frequencies on the order of a few kHz), some of the signal loss can be mitigated. Our group and others have shown promising applications of ULF MRI of human anatomy including the brain, enhanced contrast between tissues, and imaging in the presence of (and even through) metal. Although much of the required core technology has been demonstrated, ULF MRI systems still suffer from long imaging times, relatively poor quality images, and remain confined to the R and D laboratory due to the strict requirements for a low noise environment isolated from almost all ambient electromagnetic fields. Our goal in the work presented here is to move ULF MRI from a proof-of-concept in our laboratory to a functional prototype that will exploit the inherent advantages of the approach, and enable increased accessibility. Here we present results from a seven-channel SQUID-based system that achieves pre-polarization field of 100 mT over a 200 cm3 volume, is powered with all magnetic field generation from standard MRI amplifier technology, and uses off the shelf data acquisition. As our ultimate aim is unshielded operation, we also demonstrated a seven-channel system that performs ULF MRI outside of heavy magnetically-shielded enclosure. In this paper we present preliminary images and compare them to a model, and characterize the present and expected performance of this system.Let’s compare a standard 1.5-Tesla clinical MRI system with Espy et al.’s ultra-low field device. To compare quantities using the same units, I will always express the magnetic field strength in mT; a typical clinical MRI machine has a field of 1500 mT. In Section 18.3 of IPMB, Russ Hobbie and I show that the magnetization depends linearly on the magnetic field strength (Equation 18.9). The static magnetic field of Espy et al.’s machine is 0.2 mT, but for 4000 ms before spin excitation a polarization magnetic field of 100 mT is turned on. Thus, there is a difference of a factor of 15 in the magnetization, with the ultra-low device having less and the clinical machine more. Once the polarization field is turned off, the magnetic field in the ultra-low device reduces to 0.2 mT. This is only four times the earth’s magnetic field, about 0.05 mT. Espy et al. use Hemlholtz coils to cancel the earth’s field.
A magnetic field of 1500 mT is usually produced by a coil that must be kept at low temperatures to maintain the wire as a superconductor. A 100 mT field does not require superconductivity, but the needed current generates enough heat that the 510-turn copper coil needs to be cooled by liquid nitrogen, and even still the current in the coil must be turned off half the time (50% duty cycle) to avoid overheating.
Once the polarization field turns off, the spins precess with the Larmor frequency for a 0.2 mT magnetic field. The gyromagnetic ratio of protons is 42.6 kHz/mT, implying a Larmor frequency of 8.5 kHz, compared with 64,000 kHz for a clinical machine. So, the Larmor frequencies differ by a factor of 7500.
The magnetic resonance signal recorded in a clinical system is large compared to an ultra-low device because the magnetization is larger by a factor of 15 and the Larmor frequency is larger by a factor of 7500, implying a signal over a hundred thousand times larger. Espy et al. get around this problem by measuring the signal with a Superconducting Quantum Interference Device (SQUID) magnetometer, like those used in magnetoencephalography (see Section 8.9 in IPMB).
Preliminary experiments were performed in a heavy and expensive magnetically shielded room (again, like those used when measuring the MEG). However, second-order gradiometer pickup coils reduce the noise sufficiently that the shielded room is unnecessary.
To perform imaging, Espy et al. use magnetic field gradients of about 0.00025 mT/mm, compared with 0.01 mT/mm gradients typical for clinical MRI. For two objects 1 mm apart, a clinical imaging system would therefore produce a fractional frequency shift of 0.01/1500 = 0.0000067 or 6.7 ppm, whereas a low-field device has a fractional shift of 0.00025/0.2 = 0.00125 or 1250 ppm. Therefore, the clinical magnetic field needs to be extremely homogeneous (on the order of parts per million) to avoid artifacts, whereas a low-field device can function with heterogeneities hundreds of times larger.
The relaxation time constants for gray matter in the brain are reported by Espy et al. as about T1 = 600 ms and T2 = 80 ms. In clinical devices, the value of T1 is about half that, and T2 is about the same. Based on Figure 18.12 and Equation 18.35 in IPMB, I’m not surprised that T2 is largely independent of the magnetic field strength. However, in strong fields T1 increases as the square of the magnetic field (or the square of the Larmor frequency), so I was initially expecting a much smaller value of T1 for the low-field device. But once the Larmor frequency drops to values less than the typical correlation time of the spins, T1 becomes independent of the magnetic field strength (Equation 18.34 in IPMB). I assume that is what is happening here, and explains why T1 drops by only a factor of two when the magnetic field is reduced by a factor of 7500.
I find the differences between radio-frequency excitation pulses to be interesting. In clinical imaging, if the excitation pulse has a duration of about 1 ms and the Larmor frequency is 64,000 kHz, there are 64,000 oscillations of the radio-frequency magnetic field in a single π/2 pulse. Espy et al. used a 4 ms duration excitation pulse and a Larmor frequency of 8.5 kHz, implying just 34 oscillations per pulse. I have always worried that illustrations such as Figure 18.23 in IPMB mislead because they show the Larmor frequency as being not too different from the excitation pulse duration. For low-field MRI, however, this picture is realistic.
Does low-field MRI have advantages? You don’t need the heavy, expensive superconducting coil to generate a large static field, but you do need SQUID magnetometers to record the small signal, so you don’t avoid the need for cryogenics. The medicalphysicsweb article weighs the pros and cons. For instance, the power requirements for a low-field device are relatively small, and it is more portable, but the imaging times are long. The safety hazards caused by metal are much less in a low-field system, but the impact of stray magnetic fields is greater. I’m skeptical about the ultimate usefulness of ultra low-field MRI, but it’ll be fun to watch if Espy and her team can prove me wrong.
Subscribe to:
Posts (Atom)