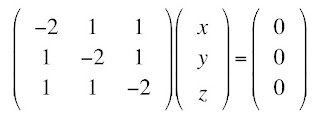|
| An image of fiber tracts in the brain using Diffusion Tensor Imaging. From: Wikipedia. |
Calculating the eigenvalues and eigenvectors of a matrix has medical and biological applications. For example, in Chapter 18 of IPMB, Russ and I discuss diffusion tensor imaging. In this technique, magnetic resonance imaging is used to measure, in each voxel, the diffusion tensor, or matrix.
This matrix is symmetric, so Dxy = Dyx, etc. It contains information about how easily spins (primarily protons in water) diffuse throughout the tissue, and about the anisotropy of the diffusion: how the rate of diffusion changes with direction. White matter in the brain is made up of bundles of nerve axons, and spins can diffuse down the long axis of an axon much easier than in the direction perpendicular to it.
Suppose you measure the diffusion matrix to be
How do you get the fiber direction from this matrix? That is the eigenvalue and eigenvector problem. Stated mathematically, the fibers are in the direction of the eigenvector corresponding to the largest eigenvalue. In other words, you can determine a coordinate system in which the diffusion matrix becomes diagonal, and the direction corresponding to the largest of the diagonal elements of the matrix is the fiber direction.
The eigenvalue problem starts with the assumption that there are some vectors r = (x, y, z) that obey the equation Dr = Dr, where D in bold is the matrix (a tensor) and D in italics is one of the eigenvalues (a scalar). We can multiply the right side by the identity matrix (1’s along the diagonal, 0’s off the diagonal) and then move this term to the left side, and get the system of equations
One obvious solution is (x, y, z) = (0, 0, 0), the trivial solution. There is a beautiful theorem from linear algebra, which I will not prove, stating that there is a nontrivial solution for (x, y, z) if and only if the determinant of the matrix is zero
I am going to assume you know how to evaluate a determinant. From this determinant, you can obtain the equation
This is a cubic equation for D, which is in general difficult to solve. However, you can show that this equation is equivalent to
Therefore, the eigenvalues of this diffusion matrix are 4, 1, and 1 (1 is a repeated eigenvalue). The largest eigenvalue is D = 4.
To find the eigenvector associated with the eigenvalue D = 4, we solve
The solution is (1, 1, 1), which points in the direction of the fibers. If you do this calculation at every voxel, you generate a fiber map of the brain, leading to beautiful pictures such as you can see at the top of this post, and here or here.
Sometimes anisotropy can be a nuisance. Suppose you just want to determine the amount of diffusion in a tissue independent of direction. You can show (see Problem 49 of Chapter 18 in IPMB) that the trace of the diffusion matrix is independent of the coordinate system. The trace is the sum of the diagonal elements of the matrix. In our example, it is 2+2+2 = 6. In the coordinate system aligned with the fiber axis, the trace is just the sum of the eigenvalues, 4+1+1 = 6 (you have to count the repeated eigenvalue twice). The trace is the same.
Now you try. Here is a new homework problem for Section 13 in Chapter 18 of IPMB.
Problem 49 1/2. Suppose the diffusion tensor in one voxel is
a) Determine the fiber direction.
b) Show explicitly in this case that the trace is the same in the original matrix as in the matrix rotated so it is diagonal.One word of warning. The examples in this blog post all happen to have simple integer eigenvalues. In general, that is not true and you need to use numerical methods to solve for the eigenvalues.
Have fun!