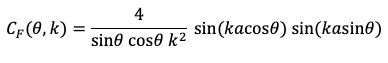In Chapter 18 of
Intermediate Physics for Medicine and Biology,
Russ Hobbie and I discuss
magnetic resonance imaging. The first step in most MRI
pulse sequences is to excite the spins in a slice of the sample. Three magnetic fields are required: A static field
B0 in the
z direction that aligns the spins, a magnetic field gradient
G in the
z direction that selects a slice with thickness Δ
z, and a
radiofrequency field in the
x direction
Bx(
t) that excites the spins.
The RF signal is called a π/2 pulse because it rotates the spins through an angle of 90 degrees (π/2
radians), from the
z direction to the
x-
y plane. The
Larmor frequency (the
angular frequency at which the spins precess at the center of the slice,
z = 0) is
ω0 =
γB0, where
γ is the
gyromagnetic ratio. Figure 18.23a of
IPMB shows the RF signal as a function of time
t: an oscillation at the Larmor frequency modulated by the
sinc function.
 |
Figure 18.23a of IPMB, showing the radiofrequency signal
used as a π/2 pulse during slice selection. |
Figure 18.24 illustrates part of the pulse sequence, showing when the RF pulse (
Bx, top) and the gradient (
G, bottom) are applied.
 |
From Figure 18.24 of IPMB, showing the part of the
pulse sequence responsible for slice selection. |
Did you ever wonder how the spins behave during this part of the pulse sequence? Do the spins flip all at once near the center of the π/2 pulse, or gradually throughout its duration? Why is there a second negative lobe of the gradient after the π/2 pulse is complete? To answer these questions, let’s analyze slice selection using the
Bloch equations—the differential equations governing how spins precess in a magnetic field. Our analysis is similar to that in Section 18.5 of
IPMB.
If we ignore
relaxation, the Bloch equations are
where
Mx,
My, and
Mz are the components of the
magnetization (a measure of the average spin orientation).
Next, let’s transform to a coordinate system (
Mx',
My',
Mz') rotating at the Larmor frequency. I will leave the details of the transformation to you (they are similar to those in Section 18.5.2 of
IPMB). We obtain
In general the Larmor frequency
γB0 is much higher than the modulation frequency
γGΔ
z/2 (thousands of oscillations occur within the central lobe of the sinc function rather than the dozen shown in Fig. 18.23a), so we average over many cycles (as in Section 18.5.3 of
IPMB). The Bloch equations simplify to
Before we can solve this system of three coupled
ordinary differential equations, we need to specify the initial conditions. At
t = −∞,
Mx' =
My' = 0 and
Mz' =
M0, where
M0 is the equilibrium value of the magnetization.
At this point, we define dimensionless variables
where
M = (
Mx,
My,
Mz). The differential equations become
where
z' ranges from -1 to 1, and the initial conditions at
t' = −∞ are
M'x' =
M'y' = 0 and
M'z' = 1.
I like this elegant system of equations, except all the primes are annoying. For the rest of this post I’ll drop the primes. Your job is to remember that these are dimensionless coordinates in a rotating coordinate system.
I wanted to solve these equations analytically, but couldn’t. I tried weird guesses involving the
sine integral and sometimes I thought I was close, but none of my attempts worked. If you, dear reader, find an analytical solution, send it to me. You will be my hero forever.
Having no analytical solution, I wrote a simple
Matlab program to solve the equations numerically.
dt=pi/1000;
T=10*pi;
nt=2*T/dt;
dz=0.05;
nz=2/dz+1;
for j=1:nz
mx(1,j)=0;
my(1,j)=0;
mz(1,j)=1;
z(j)=-1+(j-1)*dz;
end
t(1)=-T;
for i=1:nt-1
t(i+1)=t(i)+dt;
for j=1:nz
mx(i+1,j)=mx(i,j) + dt*z(j)*my(i,j);
my(i+1,j)=my(i,j) + dt*(0.5*sin(t(i))/t(i)*mz(i,j) - z(j)*mx(i,j));
mz(i+1,j)=mz(i,j) - dt*0.5*sin(t(i))/t(i)*my(i,j);
end
end
A few notes:
- I didn’t use a fancy numerical technique, just Euler’s method. To ensure accuracy, I used a tiny time step: Δt = π/1000. If you are not familiar with solving differential equations numerically, see Section 6.14 in IPMB.
- The sinc function, sin(t)/t, ranges from −∞ to +∞. I had to truncate it, so I applied the RF pulse from −10π to +10π. The middle, tall phase of the sinc function is centered at t = 0.
- I actually used the software Octave, which is a free version of Matlab. I recommend it.
My results are shown below:
Mx(
t) (blue),
My(
t) (red), and
Mz(
t) (yellow) for different values of
z, plotted for
t from −10
π to +10
π.
More notes:
- Mx is an odd function of z, whereas My and Mz are even.
- Mz changes most rapidly during the central phase of the sinc function.
- At z = 0, Mz rotates to My and then sits there constant. Remember, we are in the rotating coordinate system; in the laboratory coordinate system the spins precess at the Larmor frequency.
- As z increases, the magnetization rotates in the x-y plane for times greater than zero. The larger |z|, the higher the frequency; the local precession frequency increasingly differs from the frequency of the rotating coordinate system.
- I don’t show z = ±1. Why not? The RF pulse was designed to contain frequencies corresponding to z between -1 and 1, so z = 1 is just on the edge of the slice. What happens outside the slice? The figure below shows that the RF pulse is ineffective at exciting spins at z = 1.25 and 1.5. At exactly z = 1 the signal is just weird.
In an MRI experiment we don’t measure the magnetization at each value of
z individually, but instead detect a signal from the entire slice. Therefore, I averaged the magnetization over
z.
Mx is odd, so its average is zero. The averages of
My and
Mz are shown below.
The behavior of
My is interesting. After its central peak it decays back to zero because at all the different values of
z the spins precess at different frequencies and interfer with each other (they dephase). When averaged over the entire slice,
My is nearly zero except briefly around
t = 0.
We want
My to get big and stay big. Therefore, we need to rephase the spins after they dephase. The second lobe of the gradient field does this. All the plots so far are for times from −10
π to +10
π, and the gradient
G is on the entire time. Now, let’s extend the calculation so that for times between 10
π and 20
π the gradient is present but with opposite sign (like in Fig. 18.24) and the RF pulse is off. After time 20
π, the gradient also turns off and the only magnetic field present is
B0. The figure below shows the behavior of the spins.
We did it! When averaged over the entire slice,
My (the red trace) eventually increases, reaches one, and then stays one. It remains constant for times greater than 20
π because we ignore relaxation. The bottom line is that we found a way to excite the spins in the slice so they are all aligned in the
y direction (in the rotating coordinate system), and we are ready to apply other fancy pulse sequences to
extract information and form an image.
How did this miracle happen? The key insight is that the gradient produces an
echo, much like in
echo planar imaging.
My decays after time zero because the spins diphase, but changing the sign of the gradient rephases the spins, so they are all lined up again when we turn the gradient off at
t = 20
π. We produce an echo without using a
π pulse; it’s a
gradient echo (if you didn’t understand that last sentence, study Figures 18.31 and 18.32 in
IPMB).
Until I did this calculation, I didn’t realize that the second phase of the gradient is so crucial. I thought it corrected for a small phase shift and was only needed for high accuracy. No! The second phase produces the echo. Without it, you get no signal at all. And you have to turn it off at just the right time, or the spins will dephase again and the echo will vanish. The second lobe of the gradient is essential; the whole process fails without it.
I learned a lot from analyzing slice selection; I hope you did too.

![]()