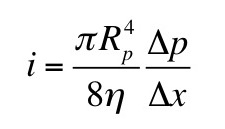Despite the compelling need mandated by the prevalence and morbidity of degenerative cartilage diseases, it is extremely difficult to study disease progression and therapeutic efficacy, either in vitro or in vivo (clinically). This is partly because no techniques have been available for nondestructively visualizing the distribution of functionally important macromolecules in living cartilage. Here we describe and validate a technique to image the glycosaminoglycan concentration ([GAG]) of human cartilage nondestructively by magnetic resonance imaging (MRI). The technique is based on the premise that the negatively charged contrast agent gadolinium diethylene triamine pentaacetic acid (Gd(DTPA)2-) will distribute in cartilage in inverse relation to the negatively charged GAG concentration. Nuclear magnetic resonance spectroscopy studies of cartilage explants demonstrated that there was an approximately linear relationship between T1 (in the presence of Gd(DTPA)2-) and [GAG] over a large range of [GAG]. Furthermore, there was a strong agreement between the [GAG] calculated from [Gd(DTPA)2-] and the actual [GAG] determined from the validated methods of calculations from [Na+] and the biochemical DMMB assay. Spatial distributions of GAG were easily observed in T1-weighted and T1-calculated MRI studies of intact human joints, with good histological correlation. Furthermore, in vivo clinical images of T1 in the presence of Gd(DTPA)2- (i.e., GAG distribution) correlated well with the validated ex vivo results after total knee replacement surgery, showing that it is feasible to monitor GAG distribution in vivo. This approach gives us the opportunity to image directly the concentration of GAG, a major and critically important macromolecule in human cartilage.
 |
| A schematic illustration of the structure of cartilage. |
Now, suppose you add a small amount of gadolinium diethylene triamine pentaacetic acid (Gd-DTPA2-); so little that you can ignore it in the equations of neutrality above. The concentrations of Gd-DTPA on the two sides of the articular surface are related by the Boltzmann factor [Gd-DTPA2-]b/[Gd-DTPA2-]t = exp(-2eV/kT) [note the factor of two in the exponent reflecting the valance -2 of Gd-DTPA], implying that [Gd-DTPA2-]b/[Gd-DTPA2-]t = ( [Na+]t/[Na+]b )2. Therefore,
We can determine [GAG-] by measuring the sodium concentration in the bath and the Gd-DTPA concentration in the bath and the tissue. Section 18.6 of IPMB describes how gadolinium shortens the T1 time constant of a magnetic resonance signal, so using T1-weighted magnetic resonance imaging you can determine the gadolinium concentration in both the bath and the tissue.
From my perspective, I like dGEMRIC because it takes two seemingly disparate parts of IPMB, the section of Donnan equilibrium and the section on how relaxation times affect magnetic resonance imaging, and combines them to create an innovative imaging method. Bashir et al.’s paper is eloquent, so I will close this blog post with their own words.
The results of this study have demonstrated that human cartilage GAG concentration can be measured and quantified in vitro in normal and degenerated tissue using magnetic resonance spectroscopy in the presence of the ionic contrast agent Gd(DTPA)2- … These spectroscopic studies therefore demonstrate the quantitative correspondence between tissue T1 in the presence of Gd(DTPA)2- and [GAG] in human cartilage. Applying the same principle in an imaging mode to obtain T1 measured on a spatially localized basis (i.e., T1-calculated images), spatial variations in [GAG] were visualized and quantified in excised intact samples…
In summary, the data presented here demonstrate the validity of the method for imaging GAG concentration in human cartilage… We now have a unique opportunity to study developmental and degenerative disease processes in cartilage and monitor the efficacy of medical and surgical therapeutic measures, for ultimately achieving a greater understanding of cartilage physiology in health and disease.